Abstract
Conventional single-dataset training often fails with new data distributions, especially in ultrasound (US) image analysis due to limited data, acoustic shadows, and speckle noise.
Therefore, constructing a universal framework for multi-heterogeneous US datasets is imperative. However, a key challenge arises: how to effectively mitigate inter-dataset interference while preserving dataset-specific discriminative features for robust downstream task?
Previous approaches utilize either a single source-specific decoder or a domain adaptation strategy, but these methods experienced a decline in performance when applied to other domains. Considering this, we propose a Universal Collaborative Mixture of Heterogeneous Source-Specific Experts (COME). Specifically, COME establishes dual structure-semantic shared experts that create a universal representation space and then collaborate with source-specific experts to extract discriminative features through providing complementary features. This design enables robust generalization by leveraging cross-datasets experience distributions and providing universal US priors for small-batch or unseen data scenarios. Extensive experiments under three evaluation modes (single-dataset, intra-organ, and inter-organ integration datasets) demonstrate COME's superiority, achieving significant mean AP improvements over state-of-the-art methods.
Details of Multiple Heterogeneous Ultrasound Dataset Integration
To develop a universal model for heterogeneous ultrasound (US) datasets, we built a benchmark of 4 breast and 4 thyroid US datasets. These datasets come from different sources and exhibit significant domain differences, such as variations in shadow artifacts, speckle noise, grayscale levels, and anatomical structures, as shown in Fig. 1
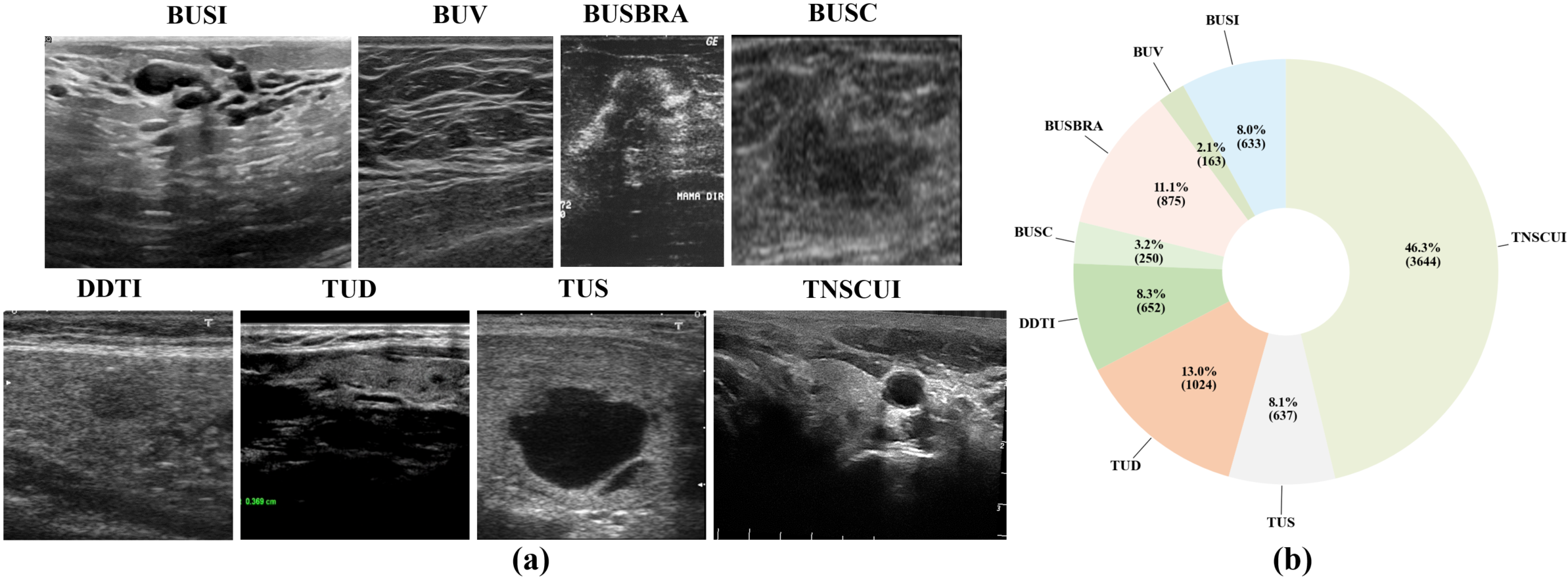
Fig. 1 (a) presents representative samples from eight datasets, highlighting their distinct imaging variations. Fig. 1 (b) illustrates the distribution of these datasets within the overall inter-organ mixed dataset, revealing a significant data imbalance.
Collaborative Mixture of Heterogeneous Source-Specific Experts (COME)
We propose the Collaborative Mixture of Heterogeneous Experts (COME), a novel framework for lesion detection across multiple heterogeneous US datasets that synergistically integrates dual structural-semantic priors with intra-dataset specific feature disentanglement, as illustrated in Fig. 2
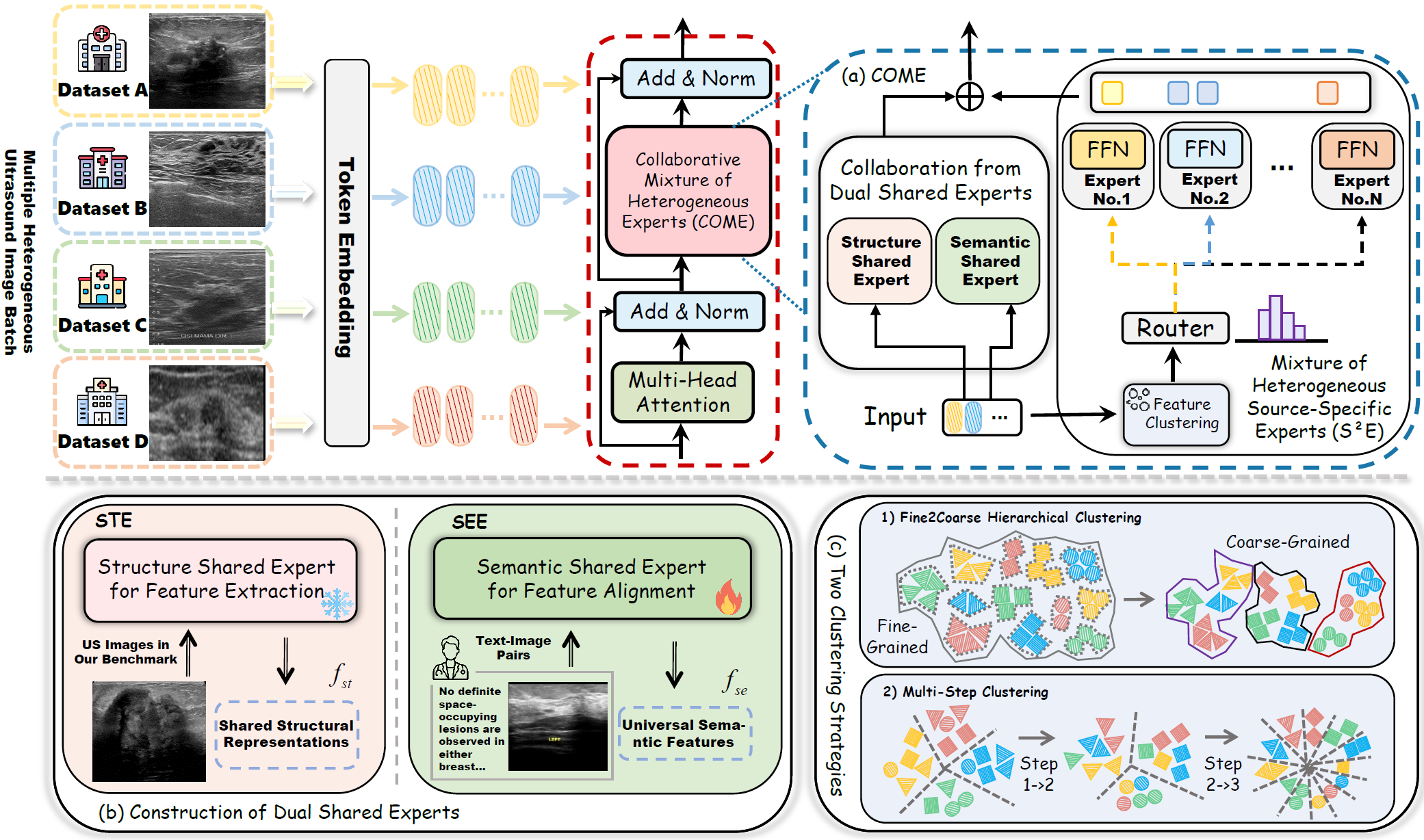
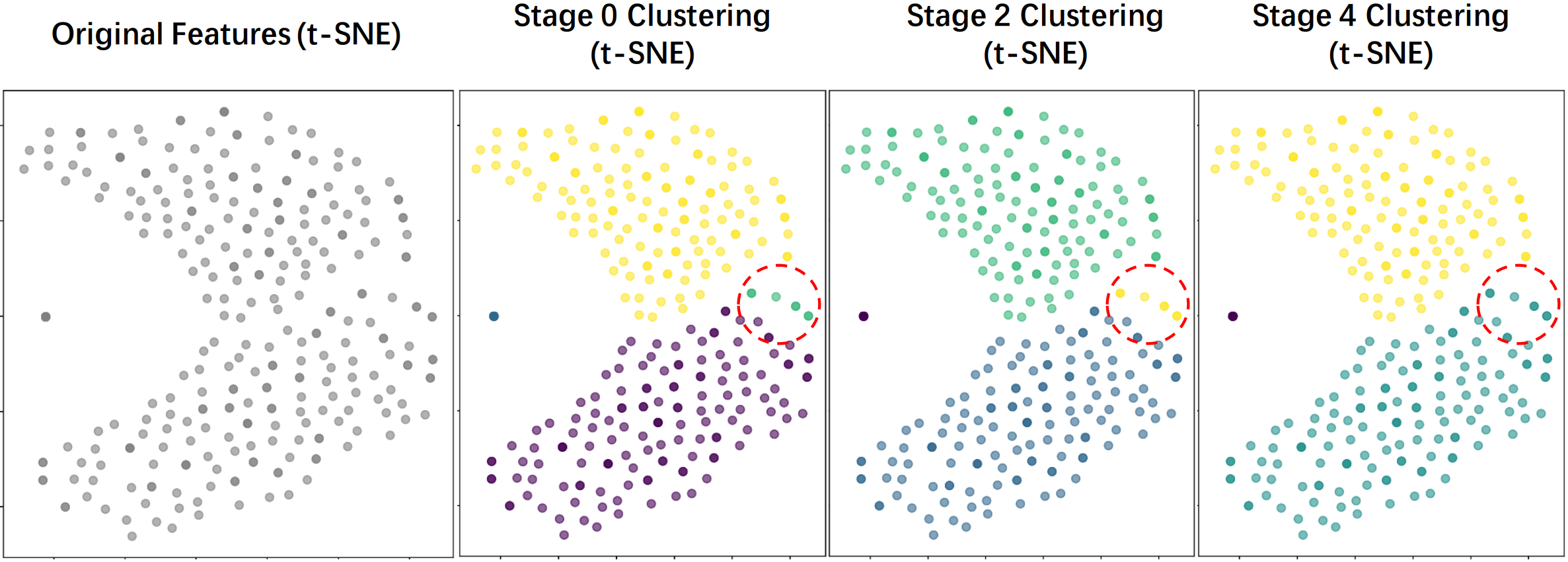
Fig. 2 (a) Overview of COME. (b) Two shared experts are constructed based on structure-semantic learning for the latent universal feature space. (c) Two clustering strategies in the S²E route tokens from the same dataset to specific experts, enabling expert specialization.
Results of Comparative Experiments
Tab. 1 compares COME with baselines across three training configurations: single-dataset, intra-organ (breast/thyroid) combinations, and inter-organ integration.
Table 1 - Lesion detection results on our benchmark, divided into top, middle, and bottom sections based on training data paradigms. We report mean AP score on each dataset. Among them, bold indicates optimal performance for three paradigms.
|
Method
|
Paradigms
|
BUSI
|
BUV
|
BUSBRA
|
BUSC
|
DDTI
|
TUD
|
TUS
|
TNSCUI
|
Mean
|
|
Open-GroundingDINO
|
Single
|
0.2495
|
0.3649
|
0.3025
|
0.3666
|
0.5391
|
0.7255
|
0.6112
|
0.6986
|
0.4822
|
|
YOLOv10
|
Single
|
0.3161
|
0.6216
|
0.5627
|
0.7052
|
0.3640
|
0.7450
|
0.4210
|
0.6907
|
0.5532
|
|
DINO
|
Single
|
0.4180
|
0.6754
|
0.5780
|
0.7313
|
0.4609
|
0.7373
|
0.6047
|
0.6945
|
0.6125
|
|
DINO-MoE
|
Single
|
0.3388
|
0.5510
|
0.5376
|
0.6706
|
0.4189
|
0.7088
|
0.5248
|
0.6669
|
0.5521
|
|
DAMEX
|
Single
|
0.3270
|
0.5653
|
0.5358
|
0.6458
|
0.4572
|
0.7101
|
0.5313
|
0.6727
|
0.5556
|
|
CerberusDet
|
Intra-Organ
|
0.3470
|
0.6980
|
0.6300
|
0.5770
|
0.5180
|
0.6780
|
0.6370
|
0.6830
|
0.5960
|
|
DINO
|
Intra-Organ
|
0.3509
|
0.6399
|
0.5193
|
0.7112
|
0.4487
|
0.6899
|
0.5666
|
0.6597
|
0.5732
|
|
DINO-MoE
|
Intra-Organ
|
0.3269
|
0.6795
|
0.5419
|
0.671
|
0.4898
|
0.7001
|
0.5801
|
0.6804
|
0.5837
|
|
DAMEX
|
Intra-Organ
|
0.3591
|
0.6990
|
0.5404
|
0.6693
|
0.5126
|
0.7000
|
0.5850
|
0.6881
|
0.5941
|
|
Our COME (Multi-Step)
|
Intra-Organ
|
0.4883
|
0.7995
|
0.6748
|
0.6879
|
0.5481
|
0.7032
|
0.5900
|
0.6875
|
0.6474
|
|
Our COME (Fine2Coarse)
|
Intra-Organ
|
0.4885
|
0.7876
|
0.6689
|
0.6434
|
0.5443
|
0.7039
|
0.5971
|
0.6922
|
0.6407
|
|
CerberusDet
|
Inter-Organ
|
0.4570
|
0.7940
|
0.6490
|
0.6300
|
0.5500
|
0.6600
|
0.6130
|
0.6620
|
0.6268
|
|
DINO
|
Inter-Organ
|
0.3825
|
0.6597
|
0.5395
|
0.7088
|
0.4583
|
0.6912
|
0.5475
|
0.6461
|
0.5792
|
|
DINO-MoE
|
Inter-Organ
|
0.4003
|
0.6838
|
0.5560
|
0.7227
|
0.5118
|
0.7103
|
0.5799
|
0.6973
|
0.6077
|
|
DAMEX
|
Inter-Organ
|
0.4098
|
0.7007
|
0.5731
|
0.7260
|
0.5214
|
0.7092
|
0.5994
|
0.6942
|
0.6167
|
|
Our COME (Multi-Step)
|
Inter-Organ
|
0.4958
|
0.7859
|
0.6912
|
0.7191
|
0.5503
|
0.7025
|
0.5972
|
0.7049
|
0.6558
|
|
Our COME (Fine2Coarse)
|
Inter-Organ
|
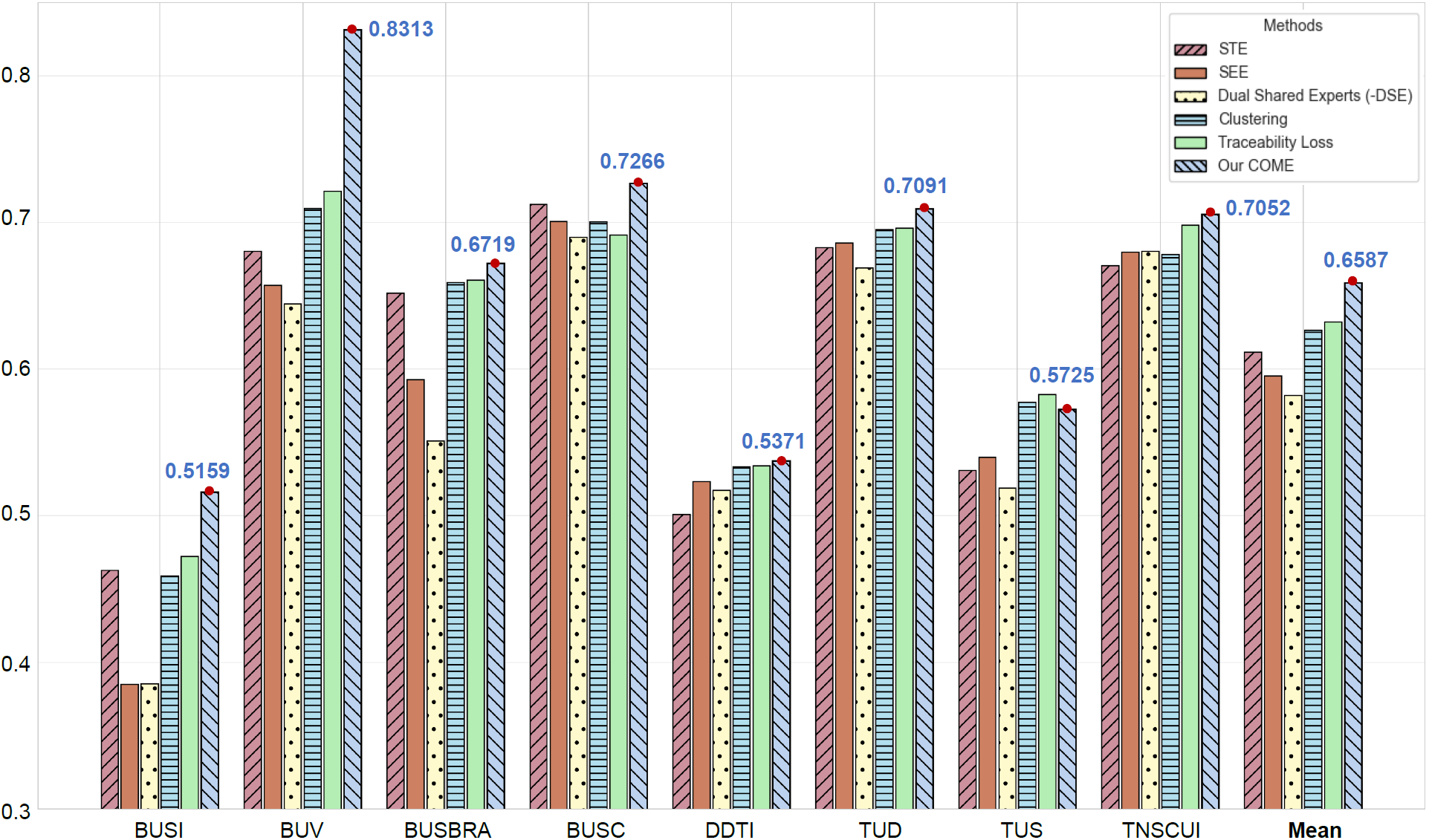
0.5159
|
0.8313
|
0.6719
|
0.7266
|
0.5371
|
0.7091
|
0.5725
|
0.7052
|
0.6587
|
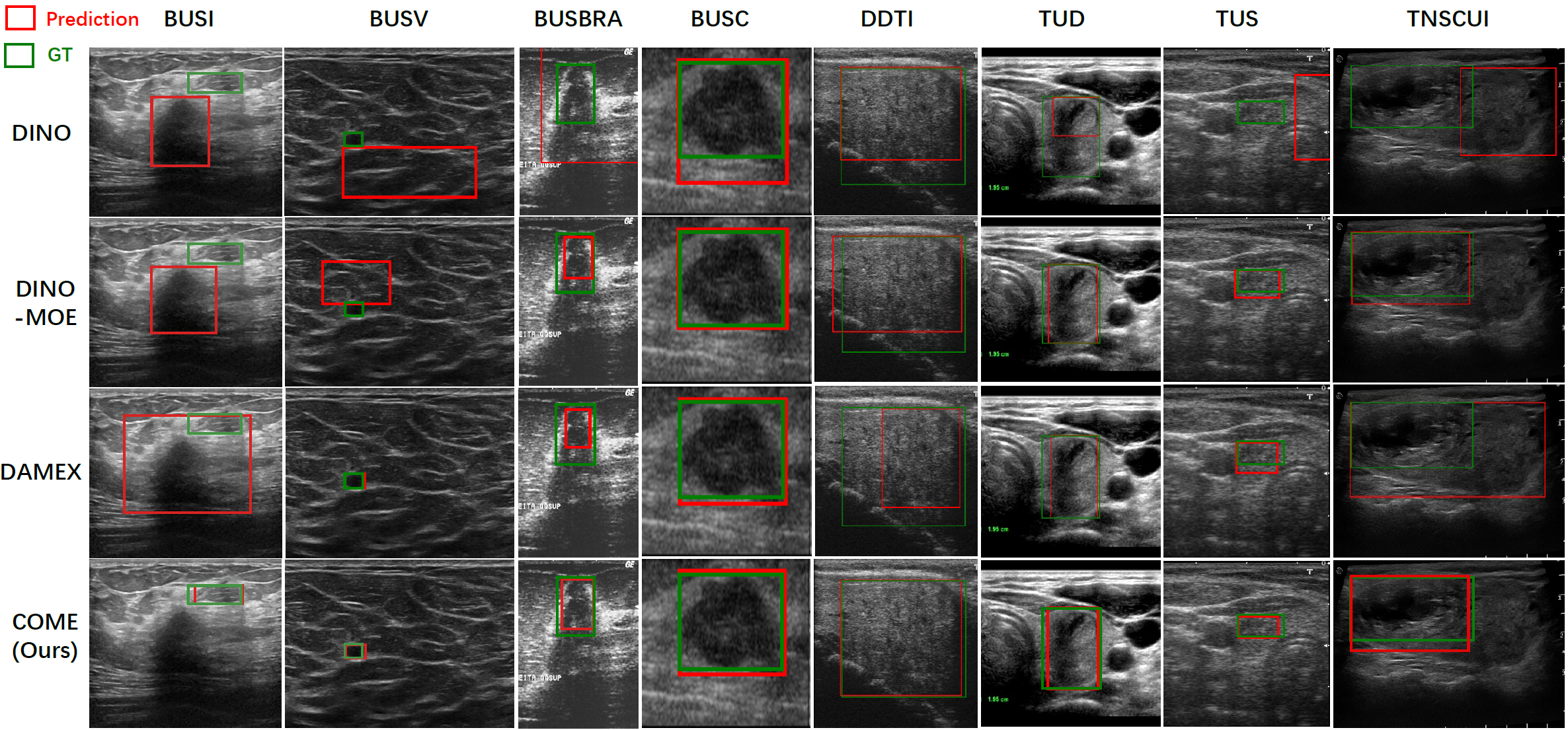
Further, we present visualization results of four models trained on the integration benchmark of eight datasets, highlighting image differences and demonstrating the superiority of our COME, as shown in Fig. 3.
Fig. 3 We visualize lesion detection on the inter-organ integration benchmark, comparing the non-MoE model DINO, the MoE-based
DINO-MoE and DAMEX, and ours. COME demonstrates strong performance across all heterogeneous datasets.
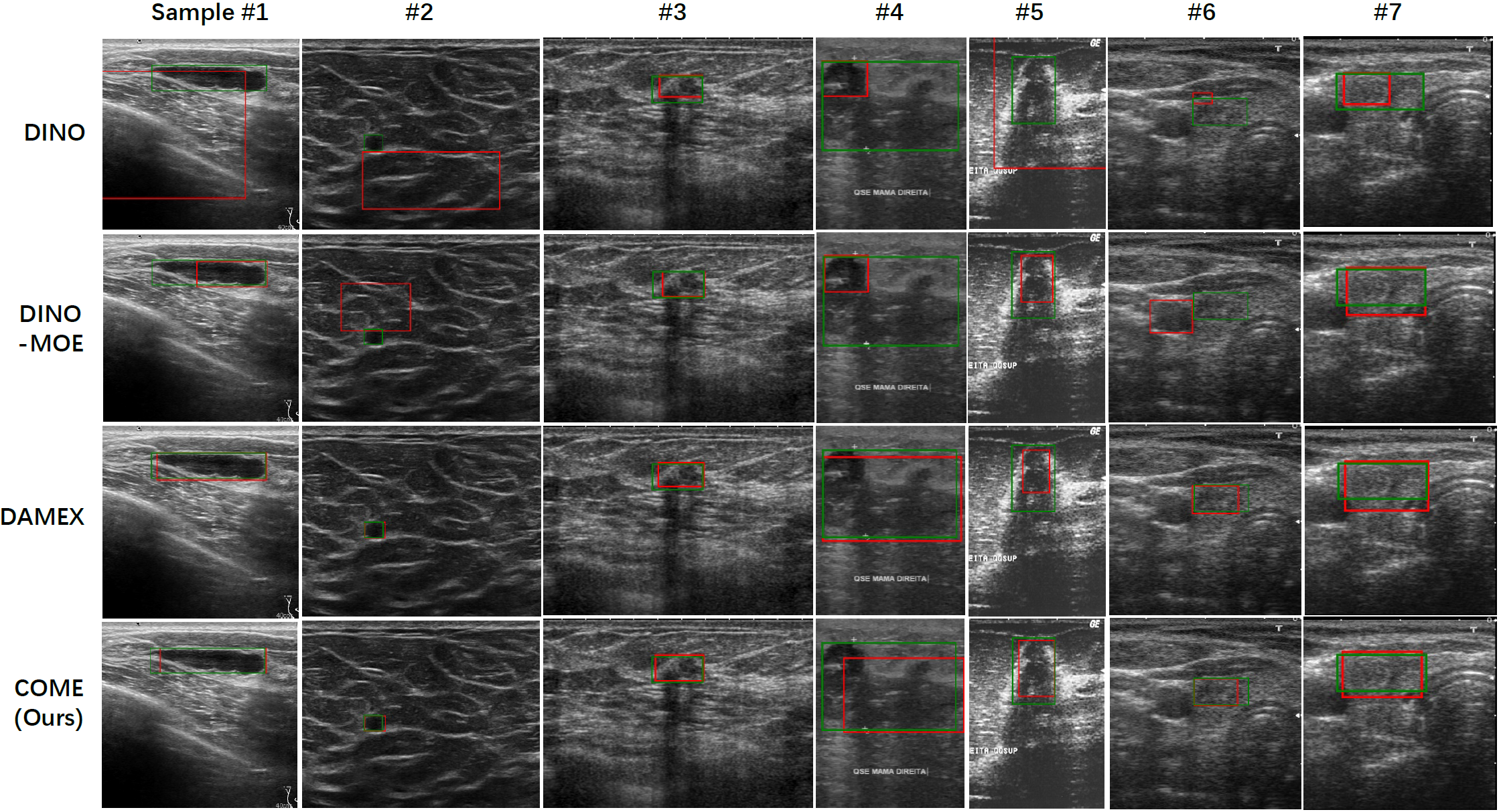
In the main text, we select one sample per dataset for comparison, as demonstrated in Fig. 3. Here, Fig. 4 shows additional lesion detection examples, demonstrating that our structure-semantic learning-based COME model delivers robust performance on diverse US images and holds promise for real-world clinical applications.
Fig. 4 Additional lesion detection examples from the inter-organ integrated dataset.